About me
Research
I am an assistant professor at the London School of Hygiene and Tropical Medicine.
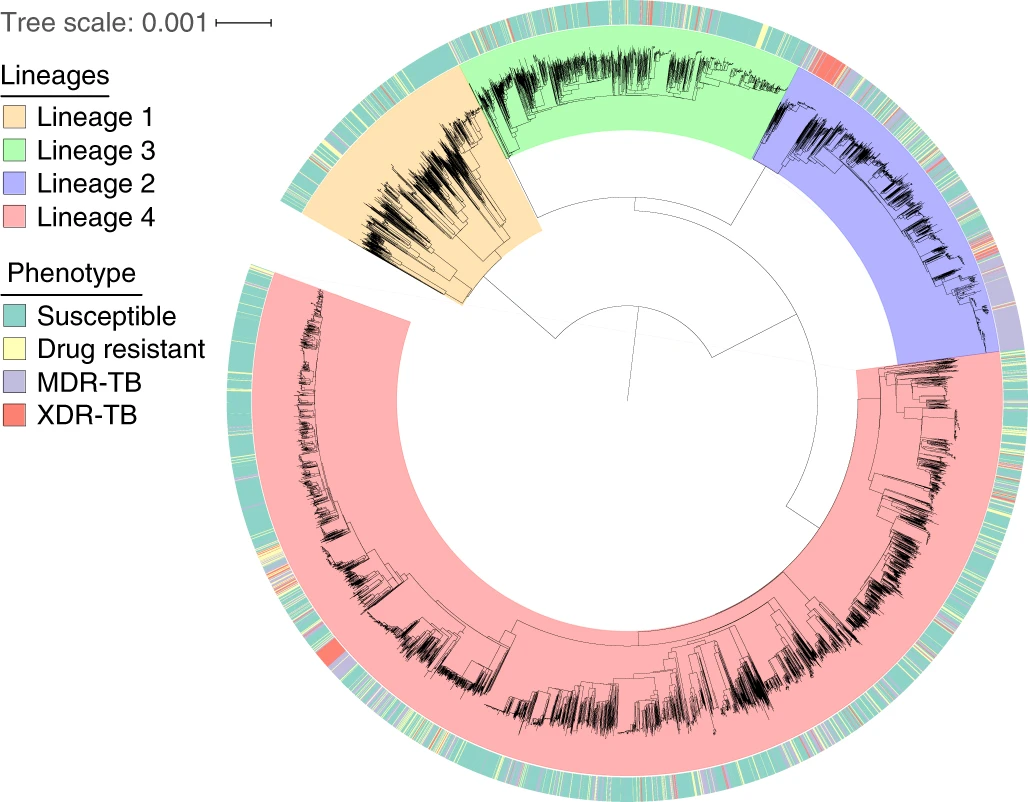
Most of my time is spent looking at the genomic diversity of Mycobacterium tuberculosis using next generation sequencing as part of the pathogenseq group.
My research entails the design and implementation of data analysis pipelines to process and collate terabytes of data from various structured and unstructured sources to provide biological insights such as population structure, transmission and drug resistance
Code
I code in
Python
R
Perl
Javascript and
html
(also learning Rust).
Here is a summary of my github commits:
Loading the data just for you.
Publications
Please check out my orcid for a complete list of my publications.